⇐ Previous topic|Next topic ⇒
Table of Contents
Paired t–test
Summary
Use the paired t–test when you have one measurement variable and two nominal variables, one of the nominal variables has only two values, and you only have one observation for each combination of the nominal variables; in other words, you have multiple pairs of observations. It tests whether the mean difference in the pairs is different from 0.
When to use it
Use the paired t–test when there is one measurement variable and two nominal variables. One of the nominal variables has only two values, so that you have multiple pairs of observations. The most common design is that one nominal variable represents individual organisms, while the other is "before" and "after" some treatment. Sometimes the pairs are spatial rather than temporal, such as left vs. right, injured limb vs. uninjured limb, etc. You can use the paired t–test for other pairs of observations; for example, you might sample an ecological measurement variable above and below a source of pollution in several streams.
As an example, volunteers count the number of breeding horseshoe crabs on beaches on Delaware Bay every year; here are data from 2011 and 2012. The measurement variable is number of horseshoe crabs, one nominal variable is 2011 vs. 2012, and the other nominal variable is the name of the beach. Each beach has one pair of observations of the measurement variable, one from 2011 and one from 2012. The biological question is whether the number of horseshoe crabs has gone up or down between 2011 and 2012.
| Beach | 2011 | 2012 | 2012−2011 |
|---|---|---|---|
| Bennetts Pier | 35282 | 21814 | -13468 |
| Big Stone | 359350 | 83500 | -275850 |
| Broadkill | 45705 | 13290 | -32415 |
| Cape Henlopen | 49005 | 30150 | -18855 |
| Fortescue | 68978 | 125190 | 56212 |
| Fowler | 8700 | 4620 | -4080 |
| Gandys | 18780 | 88926 | 70146 |
| Higbees | 13622 | 1205 | -12417 |
| Highs | 24936 | 29800 | 4864 |
| Kimbles | 17620 | 53640 | 36020 |
| Kitts Hummock | 117360 | 68400 | -48960 |
| Norburys Landing | 102425 | 74552 | -27873 |
| North Bowers | 59566 | 36790 | -22776 |
| North Cape May | 32610 | 4350 | -28260 |
| Pickering | 137250 | 110550 | -26700 |
| Pierces Point | 38003 | 43435 | 5432 |
| Primehook | 101300 | 20580 | -80720 |
| Reeds | 62179 | 81503 | 19324 |
| Slaughter | 203070 | 53940 | -149130 |
| South Bowers | 135309 | 87055 | -48254 |
| South CSL | 150656 | 112266 | -38390 |
| Ted Harvey | 115090 | 90670 | -24420 |
| Townbank | 44022 | 21942 | -22080 |
| Villas | 56260 | 32140 | -24120 |
| Woodland | 125 | 1260 | 1135 |
As you might expect, there's a lot of variation from one beach to the next. If the difference between years is small relative to the variation within years, it would take a very large sample size to get a significant two-sample t–test comparing the means of the two years. A paired t–test just looks at the differences, so if the two sets of measurements are correlated with each other, the paired t–test will be more powerful than a two-sample t–test. For the horseshoe crabs, the P value for a two-sample t–test is 0.110, while the paired t–test gives a P value of 0.045.
You can only use the paired t–test when there is just one observation for each combination of the nominal values. If you have more than one observation for each combination, you have to use two-way anova with replication. For example, if you had multiple counts of horseshoe crabs at each beach in each year, you'd have to do the two-way anova.
You can only use the paired t–test when the data are in pairs. If you wanted to compare horseshoe crab abundance in 2010, 2011, and 2012, you'd have to do a two-way anova without replication.
"Paired t–test" is just a different name for "two-way anova without replication, where one nominal variable has just two values"; the results are mathematically identical. The paired design is a common one, and if all you're doing is paired designs, you should call your test the paired t–test; it will sound familiar to more people. But if some of your data sets are in pairs, and some are in sets of three or more, you should call all of your tests two-way anovas; otherwise people will think you're using two different tests.
Null hypothesis
The null hypothesis is that the mean difference between paired observations is zero. When the mean difference is zero, the means of the two groups must also be equal. Because of the paired design of the data, the null hypothesis of a paired t–test is usually expressed in terms of the mean difference.
Assumption
The paired t–test assumes that the differences between pairs are normally distributed; you can use the histogram spreadsheet described on that page to check the normality. If the differences between pairs are severely non-normal, it would be better to use the Wilcoxon signed-rank test. I don't think the test is very sensitive to deviations from normality, so unless the deviation from normality is really obvious, you shouldn't worry about it.
The paired t–test does not assume that observations within each group are normal, only that the differences are normal. And it does not assume that the groups are homoscedastic.
How the test works
The first step in a paired t–test is to calculate the difference for each pair, as shown in the last column above. Then you use a one-sample t–test to compare the mean difference to 0. So the paired t–test is really just one application of the one-sample t–test, but because the paired experimental design is so common, it gets a separate name.
Examples
Wiebe and Bortolotti (2002) examined color in the tail feathers of northern flickers. Some of the birds had one "odd" feather that was different in color or length from the rest of the tail feathers, presumably because it was regrown after being lost. They measured the yellowness of one odd feather on each of 16 birds and compared it with the yellowness of one typical feather from the same bird. There are two nominal variables, type of feather (typical or odd) and the individual bird, and one measurement variable, yellowness. Because these birds were from a hybrid zone between red-shafted flickers and yellow-shafted flickers, there was a lot of variation among birds in color, making a paired analysis more appropriate. The difference was significant (P=0.001), with the odd feathers significantly less yellow than the typical feathers (higher numbers are more yellow).
| Yellowness index | ||
|---|---|---|
| Bird | Typical feather | Odd feather |
| A | -0.255 | -0.324 |
| B | -0.213 | -0.185 |
| C | -0.19 | -0.299 |
| D | -0.185 | -0.144 |
| E | -0.045 | -0.027 |
| F | -0.025 | -0.039 |
| G | -0.015 | -0.264 |
| H | 0.003 | -0.077 |
| I | 0.015 | -0.017 |
| J | 0.02 | -0.169 |
| K | 0.023 | -0.096 |
| L | 0.04 | -0.33 |
| M | 0.04 | -0.346 |
| N | 0.05 | -0.191 |
| O | 0.055 | -0.128 |
| P | 0.058 | -0.182 |
Wilder and Rypstra (2004) tested the effect of praying mantis excrement on the behavior of wolf spiders. They put 12 wolf spiders in individual containers; each container had two semicircles of filter paper, one semicircle that had been smeared with praying mantis excrement and one without excrement. They observed each spider for one hour, and measured its walking speed while it was on each half of the container. There are two nominal variables, filter paper type (with or without excrement) and the individual spider, and one measurement variable (walking speed). Different spiders may have different overall walking speed, so a paired analysis is appropriate to test whether the presence of praying mantis excrement changes the walking speed of a spider. The mean difference in walking speed is almost, but not quite, significantly different from 0 (t=2.11, 11 d.f., P=0.053).
Graphing the results
If there are a moderate number of pairs, you could either plot each individual value on a bar graph, or plot the differences. Here is one graph in each format for the flicker data:
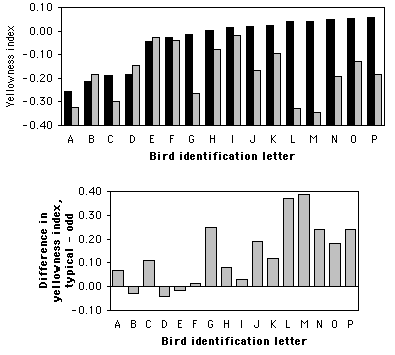
Related tests
The paired t–test is mathematically equivalent to one of the hypothesis tests of a two-way anova without replication. The paired t–test is simpler to perform and may sound familiar to more people. You should use two-way anova if you're interested in testing both null hypotheses (equality of means of the two treatments and equality of means of the individuals); for the horseshoe crab example, if you wanted to see whether there was variation among beaches in horseshoe crab density, you'd use two-way anova and look at both hypothesis tests. In a paired t–test, the means of individuals are so likely to be different that there's no point in testing them.
If you have multiple observations for each combination of the nominal variables (such as multiple observations of horseshoe crabs on each beach in each year), you have to use two-way anova with replication.
If you ignored the pairing of the data, you would use a one-way anova or a two-sample t–test. When the difference of each pair is small compared to the variation among pairs, a paired t–test can give you a lot more statistical power than a two-sample t–test, so you should use the paired test whenever your data are in pairs.
One non-parametric analogue of the paired t–test is Wilcoxon signed-rank test; you should use if the differences are severely non-normal. A simpler and even less powerful test is the sign test, which considers only the direction of difference between pairs of observations, not the size of the difference.
How to do the test
Spreadsheet
Spreadsheets have a built-in function to perform paired t–tests. Put the "before" numbers in one column, and the "after" numbers in the adjacent column, with the before and after observations from each individual on the same row. Then enter =TTEST(array1, array2, tails, type), where array1 is the first column of data, array2 is the second column of data, tails is normally set to 2 for a two-tailed test, and type is set to 1 for a paired t–test. The result of this function is the P value of the paired t–test.
Even though it's easy to do yourself, I've written a spreadsheet to do a paired t-test.
Web pages
There are web pages to do paired t–tests here, here, here, and here.
R
Salvatore Mangiafico's R Companion has a sample R program for the paired t–test.
SAS
To do a paired t–test in SAS, you use PROC TTEST with the PAIRED option. Here is an example using the feather data from above:
DATA feathers; INPUT bird $ typical odd; DATALINES; A -0.255 -0.324 B -0.213 -0.185 C -0.190 -0.299 D -0.185 -0.144 E -0.045 -0.027 F -0.025 -0.039 G -0.015 -0.264 H 0.003 -0.077 I 0.015 -0.017 J 0.020 -0.169 K 0.023 -0.096 L 0.040 -0.330 M 0.040 -0.346 N 0.050 -0.191 O 0.055 -0.128 P 0.058 -0.182 ; PROC TTEST DATA=feathers; PAIRED typical*odd; RUN;
The results include the following, which shows that the P value is 0.0010:
t–tests
Difference DF t Value Pr > |t|
typical - odd 15 4.06 0.0010
Power analysis
To estimate the sample sizes needed to detect a mean difference that is significantly different from zero, you need the following:
- the effect size, or the mean difference. In the feather data used above, the mean difference between typical and odd feathers is 0.137 yellowness units.
- the standard deviation of differences. Note that this is not the standard deviation within each group. For example, in the feather data, the standard deviation of the differences is 0.135; this is not the standard deviation among typical feathers, or the standard deviation among odd feathers, but the standard deviation of the differences;
- alpha, or the significance level (usually 0.05);
- power, the probability of rejecting the null hypothesis when it is false and the true difference is equal to the effect size (0.80 and 0.90 are common values).
As an example, let's say you want to do a study comparing the redness of typical and odd tail feathers in cardinals. The closest you can find to preliminary data is the Weibe and Bortolotti (2002) paper on yellowness in flickers. They found a mean difference of 0.137 yellowness units, with a standard deviation of 0.135; you arbitrarily decide you want to be able to detect a mean difference of 0.10 redness units in your cardinals. In G*Power, choose "t tests" under Test Family and "Means: Difference between two dependent means (matched pairs)" under Statistical Test. Choose "A priori: Compute required sample size" under Type of Power Analysis. Under Input Parameters, choose the number of tails (almost always two), the alpha (usually 0.05), and the power (usually something like 0.8 or 0.9). Click on the "Determine" button and enter the effect size you want (0.10 for our example) and the standard deviation of differences, then hit the "Calculate and transfer to main window" button. The result for our example is a total sample size of 22, meaning that if the true mean difference is 0.10 redness units and the standard deviation of differences is 0.135, you'd have a 90% chance of getting a result that's significant at the P<0.05 level if you sampled typical and odd feathers from 22 cardinals.
References
Wiebe, K.L., and G.R. Bortolotti. 2002. Variation in carotenoid-based color in northern flickers in a hybrid zone. Wilson Bulletin 114: 393-400.
Wilder, S.M., and A.L. Rypstra. 2004. Chemical cues from an introduced predator (Mantodea, Mantidae) reduce the movement and foraging of a native wolf spider (Araneae, Lycosidae) in the laboratory. Environmental Entomology 33: 1032-1036.
⇐ Previous topic|Next topic ⇒
Table of Contents
This page was last revised July 20, 2015. Its address is http://www.biostathandbook.com/pairedttest.html. It may be cited as:
McDonald, J.H. 2014. Handbook of Biological Statistics (3rd ed.). Sparky House Publishing, Baltimore, Maryland. This web page contains the content of pages 180-185 in the printed version.
©2014 by John H. McDonald. You can probably do what you want with this content; see the permissions page for details.

